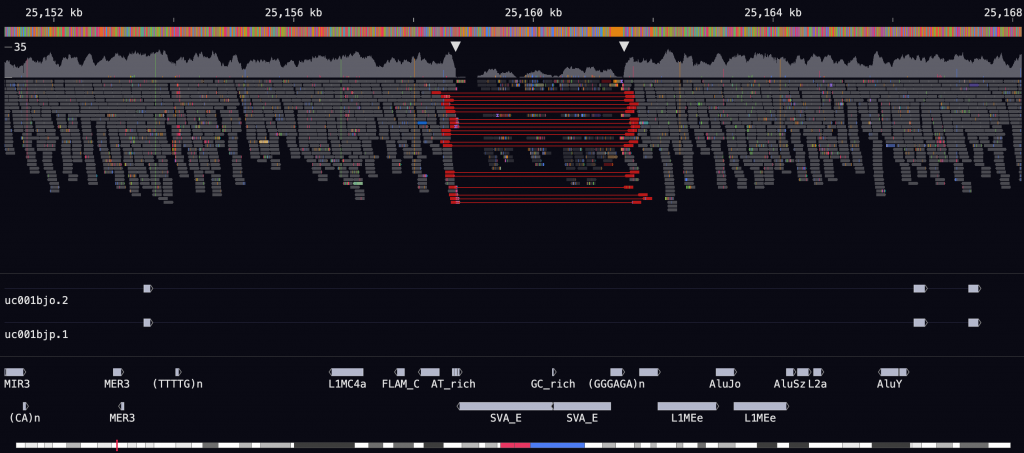
WCRC researcher Dr Kez Cleal and his team have unveiled a groundbreaking tool called Genome-Wide (GW), a cutting-edge software designed to transform how scientists visualise and analyse genomic data.
Detailed in a recent pre-print on bioRxiv titled “GW: Ultra-Fast Chromosome-Scale Visualisation of Genomics Data,” the software is compared to a “Google Earth” for genomics, allowing researchers to explore massive datasets with unprecedented speed.
GW boasts a processing speed over 100 times faster than existing tools, offering scientists a major advantage in analysing complex genetic information. This breakthrough is particularly important in cancer research, where understanding large-scale structural changes in the genome is key to unlocking the mechanisms behind the disease.
Dr Cleal said: “With GW, we’re able to dynamically visualise genome-scale changes, giving us the ability to explore and understand complex genetic structures in ways that were not possible before. This opens the door to more efficient cancer research, helping us to quickly identify potential diagnostic markers and therapeutic targets.”
The GW software enables real-time observation of these changes, speeding up crucial research tasks. Dr Cleale’s team is now focusing on two major goals: developing diagnostic tools to help identify and categorise patients with abnormal genomic structures, and investigating the biological mechanisms driving these changes. Their ultimate aim is to explore new ways to target these processes for cancer treatment.
This innovation could significantly advance the study of cancer and improve how researchers approach diagnosis and potential therapies.